7.4 Database Integration: Air Quality, Mortality, and Environmental Justice Data
The development of this training module was led by Cavin Ward-Caviness with contributions from Alexis Payton.
Disclaimer: The views expressed in this document are those of the author and do not necessarily reflect the views or policies of the U.S. EPA.
Introduction to Training Module
This training module provides an example analysis based on the integration of data across multiple environmental health databases. This module specifically guides trainees through an explanation of how the data were downloaded and organized, and then details the loading of required packages and datasets. Then, code is provided for visualizing county-level air pollution measures, including PM2.5, NO2, and SO2. These measures were obtained through U.S. EPA monitoring stations are visualized here as the yearly average. Air pollution concentrations are then evaluated for potential relationship to the health outcome, mortality. Specifically, age-adjusted mortality rates are organized and statistically related to PM2.5 concentrations through linear regression modeling. Crude statistical models are first provided that do not take into account the influence of potential confounders. Then, statistical models are used that adjust for potential confounders, including adult smoking rates, obesity, food environment indicators, physical activity, employment status, rural vs urban living percentages, sex, ethnicity, and race. Results from these models point to the finding that areas with higher percentages of African-Americans may be experiencing higher impacts from PM2.5 on mortality. This relationship is of high interest, as it represents a potential environmental justice issue.
Introduction to Exposure and Health Databases
In this training module, we will use publicly available exposure and health databases to examine associations between air quality and mortality across the entire U.S. Specific databases that we will query include the following:
EPA Air Quality data: As an example air pollutant exposure dataset, 2016 annual average data from the EPA Air Quality System database will be analyzed, using data downloaded and organized from the following website: https://aqs.epa.gov/aqsweb/airdata/annual_conc_by_monitor_2016.zip
CDC Health Outcome data: As an example health outcome dataset, the 2016 CDC Underlying Cause of Death dataset, from the WONDER (Wide-ranging ONline Data for Epidemiologic Research) website will be analyzed, using All-Cause Mortality Rates downloaded and organized from the following website: https://wonder.cdc.gov/ucd-icd10.html
Human covariate data: The potential influence of covariates (e.g., race) and other confounders will be analyzed using data downloaded and organized from the following 2016 county-level resource: https://www.countyhealthrankings.org/explore-health-rankings/rankings-data-documentation/national-data-documentation-2010-2019
Training Module’s Environmental Health Questions
This training module was specifically developed to answer the following environmental health questions:
- What areas of the U.S. are most heavily monitored for air quality?
- Is there an association between long-term, ambient PM2.5 concentrations and mortality at the county level? Stated another way we are asking: Do counties with higher annual average PM2.5 concentrations also have higher all-cause mortality rates?
- What is the difference when running crude statistical models vs. statistical models that adjust for potential confounding, when evaluating the relationship between PM2.5 and mortality?
- Do observed associations differ when comparing between counties with a higher vs. lower percentage of African-Americans which can indicate environmental justice concerns?
Script Preparations
Installing required R packages
If you already have these packages installed, you can skip this step, or you can run the below code which checks installation status for you
Set your working directory
Let’s start by loading the datasets needed for this training module. As detailed in the introduction, these data were previously downloaded and organized, and specifically made available for this training exercise as a compiled RDataset, containing organized dataframes ready to analyze.
We can now read in these organized data using the load() function.
First let’s take a look at the geographic data, starting with the county-level shapefile (counties_shapefile). This dataframe contains the location information for the counties which we will use for visualizations.
head(counties_shapefile)
#> Simple feature collection with 6 features and 9 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -102.042 ymin: 37.38839 xmax: -84.79633 ymax: 43.49961
#> Geodetic CRS: NAD83
#> STATEFP COUNTYFP COUNTYNS AFFGEOID GEOID NAME LSAD ALAND AWATER
#> 1 19 107 00465242 0500000US19107 19107 Keokuk 06 1500067253 1929323
#> 2 19 189 00465283 0500000US19189 19189 Winnebago 06 1037261946 3182052
#> 3 20 093 00485011 0500000US20093 20093 Kearny 06 2254696689 1133601
#> 4 20 123 00485026 0500000US20123 20123 Mitchell 06 1817632928 44979981
#> 5 20 187 00485055 0500000US20187 20187 Stanton 06 1762104518 178555
#> 6 21 005 00516849 0500000US21005 21005 Anderson 06 522745702 6311537
#> geometry
#> 1 MULTIPOLYGON (((-92.41199 4...
#> 2 MULTIPOLYGON (((-93.97076 4...
#> 3 MULTIPOLYGON (((-101.5419 3...
#> 4 MULTIPOLYGON (((-98.49007 3...
#> 5 MULTIPOLYGON (((-102.0419 3...
#> 6 MULTIPOLYGON (((-85.16919 3...These geographic data are represented by the following columns. (Some columns are not described below, since we don’t need them for these analyses.):
STATEFP: State FIPS code (1 or 2 digits)COUNTYFP: County FIPS Code (3 digits)GEOID: Geographic identifier that combines the state and county FIPS codesNAME: County namegeometry: Latitude and longitude coordinates
Now let’s view the EPA Air Quality Survey (AQS) data (epa_ap_county) collected from 2016. This dataframe represents county-level air quality measures, as detailed above. This dataframe is in melted (or long) format, meaning that different air quality measures are organized across rows, with variable measure indicators in the Parameter.Name, Units.of.Measure, and County_Avg columns.
head(epa_ap_county)
#> State.Code State.Name County.Code County.Name State_County_Code Parameter.Name
#> 1 1 Alabama 3 Baldwin 1003 PM25
#> 2 1 Alabama 27 Clay 1027 PM25
#> 3 1 Alabama 33 Colbert 1033 PM25
#> 4 1 Alabama 49 DeKalb 1049 PM25
#> 5 1 Alabama 55 Etowah 1055 PM25
#> 6 1 Alabama 69 Houston 1069 PM25
#> Units.of.Measure County_Avg
#> 1 Micrograms/cubic meter (LC) 7.226446
#> 2 Micrograms/cubic meter (LC) 7.363793
#> 3 Micrograms/cubic meter (LC) 7.492241
#> 4 Micrograms/cubic meter (LC) 7.695690
#> 5 Micrograms/cubic meter (LC) 8.195575
#> 6 Micrograms/cubic meter (LC) 7.061538These air quality data are represented by the following columns:
State.Code: State FIPS code (1 or 2 digits)State.Name: State nameCounty.Code: County FIPS code (1-3 digits)County.Name: County nameState_County_Code: Combined state and county code (separated by a 0)Parameter.Name: Name of the air pollutantUnits.of.Measure: Units of measurementCounty_Avg: County average
These data can be restructured to view air quality measures as separate variables labeled across columns using:
# transform from the "long" to "wide" format for the pollutants
epa_ap_county <- epa_ap_county %>%
select(-Units.of.Measure) %>%
unique() %>%
tidyr::spread(Parameter.Name, County_Avg)
head(epa_ap_county)
#> State.Code State.Name County.Code County.Name State_County_Code NO2 PM25 SO2
#> 1 1 Alabama 3 Baldwin 1003 NA 7.226446 NA
#> 2 1 Alabama 27 Clay 1027 NA 7.363793 NA
#> 3 1 Alabama 33 Colbert 1033 NA 7.492241 NA
#> 4 1 Alabama 49 DeKalb 1049 NA 7.695690 NA
#> 5 1 Alabama 55 Etowah 1055 NA 8.195575 NA
#> 6 1 Alabama 69 Houston 1069 NA 7.061538 NANote that we can now see the specific pollutant variables NO2, PM25, and SO2 on the far right.
Population-Weighted vs. Unweighted Exposures
Here we pause briefly to speak on population-weighted vs unweighted exposures. The analysis we will be undertaking is known as an “ecological” analysis where we are looking at associations by area, e.g. county. When studying environmental exposures by area a common practice is to try to weight the exposures by the population so that exposures better represent the “burden” faced by the population. Ideally for this you would want a systematic model or assessment of the exposure that corresponded with a fine-scale population estimate so that for each county you could weight exposures within different areas of the county by the population exposed. This sparse monitor data (we will examine the population covered later in the tutorial) is not population weighted, but should you see similar analyses with population weighting of exposures you should simply be aware that this better captures the “burden” of exposure experienced by the population within the area estimated, typically zip code or county.
Now let’s view the CDC’s mortality dataset collected from 2016 (cdc_mortality):
head(cdc_mortality)
#> Notes County County.Code Deaths Population Crude.Rate Age.Adjusted.Rate
#> 1 NA Autauga County, AL 1001 520 55416 938.36 884.40
#> 2 NA Baldwin County, AL 1003 1974 208563 946.48 716.92
#> 3 NA Barbour County, AL 1005 256 25965 985.94 800.68
#> 4 NA Bibb County, AL 1007 239 22643 1055.51 927.67
#> 5 NA Blount County, AL 1009 697 57704 1207.89 989.37
#> 6 NA Bullock County, AL 1011 133 10362 1283.54 1063.00
#> Age.Adjusted.Rate.Standard.Error
#> 1 39.46
#> 2 16.58
#> 3 51.09
#> 4 61.03
#> 5 38.35
#> 6 93.69These mortality data are represented by the following columns. We’ll just ignore the Notes column for our purposes:
County: County name with the state abbreviationCounty.Code: Combined state and county code (separated by a 0)Deaths: Number of deaths in 2016Population: County population in 2016Crude.Rate: Death rate (\(\frac{Number~of~Deaths}{Population}* 100,000\))Age.Adjusted.Rate: age-adjusted death rate (\(\sum{(Age~Specific~Death~Rate * Standard~Population~Weight)} * 100,000\))Age.Adjusted.Rate.Standard.Error: Standard error of the age-adjusted rate
We can create a visualization of the age-adjusted death rate and air pollutants throughout the U.S. to further inform what these data look like:
# Can merge them by the FIPS county code which we need to create for the counties_shapefile
counties_shapefile$State_County_Code <- as.character(as.numeric(paste0(counties_shapefile$STATEFP, counties_shapefile$COUNTYFP)))
# Let's merge in the air pollution and mortality data and plot it
counties_shapefile <- merge(counties_shapefile, epa_ap_county, by.x = "State_County_Code", by.y = "State_County_Code", all.x=TRUE)
counties_shapefile <- merge(counties_shapefile, cdc_mortality, by.x = "State_County_Code", by.y = "County.Code")
# Will remove alaska and hawaii just so we can look at the continental USA
county_plot <- subset(counties_shapefile, !STATEFP %in% c("02","15"))
# We can start with a simple plot of age-adjusted mortality rate, PM2.5, NO2, and SO2 levels across the U.S.
plot(county_plot[,c("Age.Adjusted.Rate","PM25","NO2","SO2")])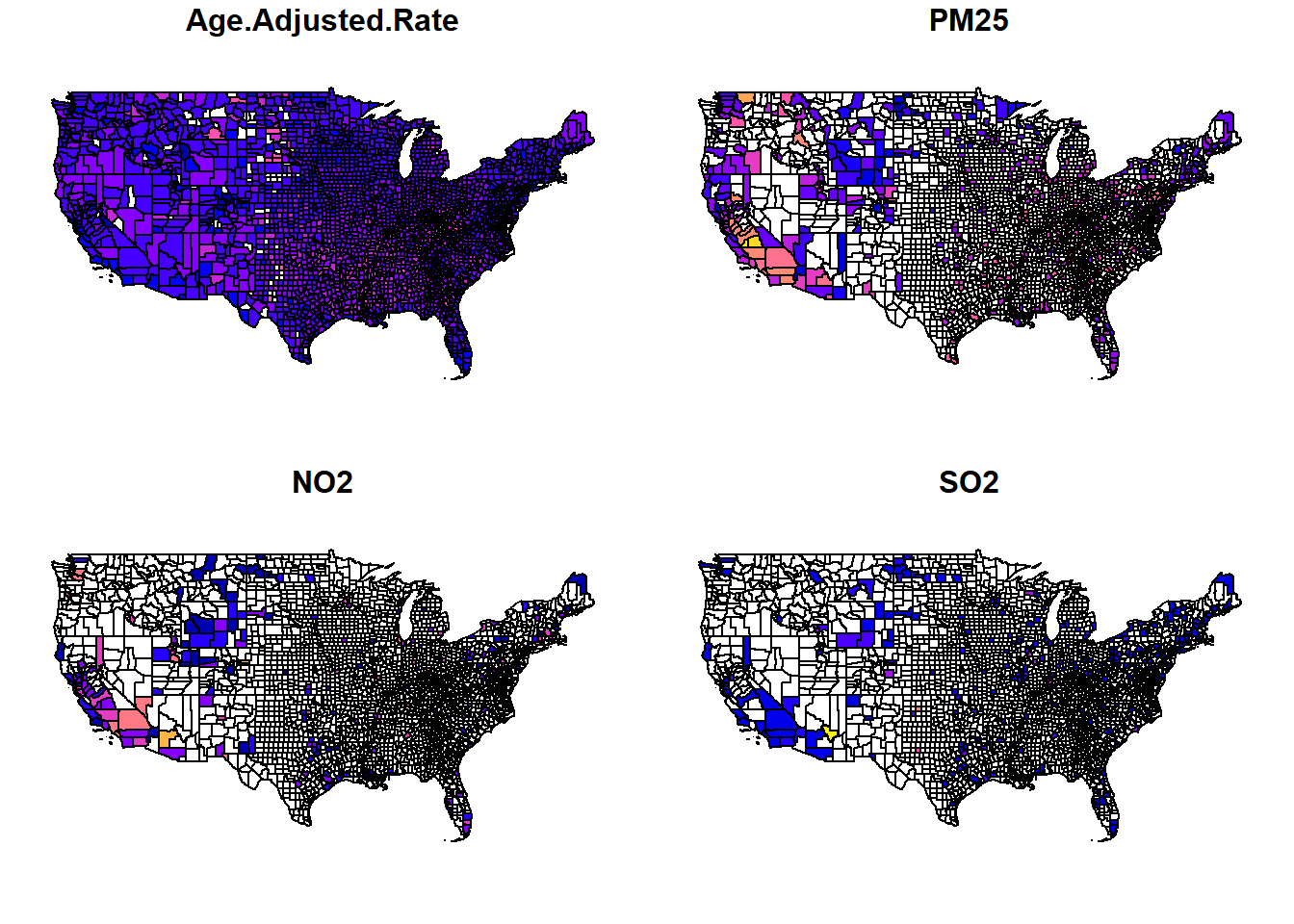
You can see that these result in the generation of four different nationwide plots, showing the distributions of age-adjusted mortality rates, PM2.5 concentrations, NO2 concentrations, and SO2 concentrations, averaged per county.
Let’s make a nicer looking plot with ggplot(), looking just at PM2.5 levels:
ggplot(data = county_plot) +
geom_sf(aes(fill = PM25)) +
# Changing colors
scale_fill_viridis_c(option ="plasma", name ="PM2.5",
guide = guide_colorbar(
direction = "horizontal",
barheight = unit(2, units = "mm"),
barwidth = unit(50, units = "mm"),
draw.ulim = F,
title.position = 'top',
# some shifting around
title.hjust = 0.5,
label.hjust = 0.5)) +
ggtitle(expression(2016~Annual~PM[2.5]~EPA~Monitors)) +
theme_map() +
theme(plot.title = element_text(hjust = 0.5, size = 22)) 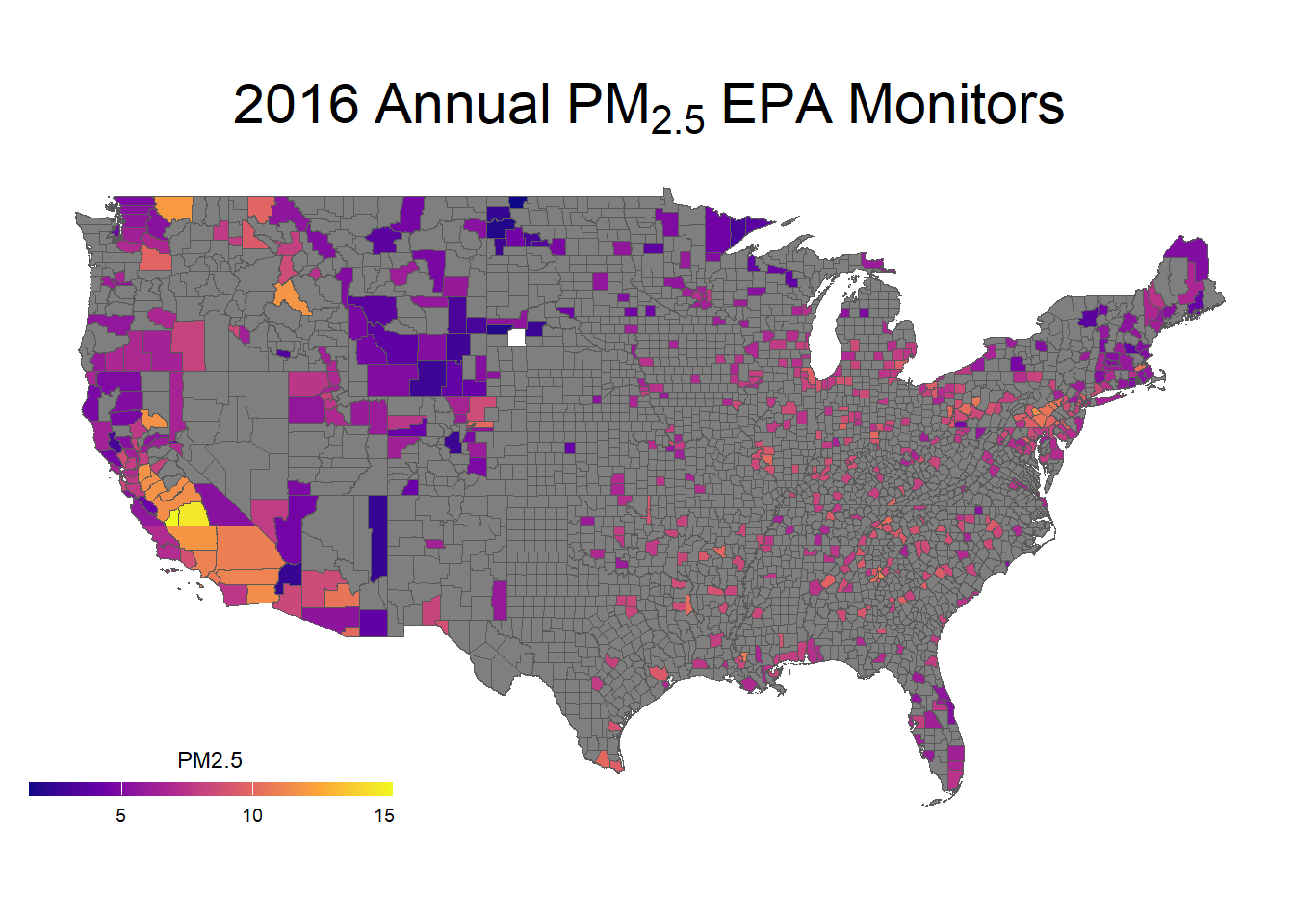
Answer to Environmental Health Question 1
With this, we can answer Environmental Health Question #1: What areas of the U.S. are most heavily monitored for air quality?
Answer: We can tell from the PM2.5 specific plot that air monitors are densely located in California, and other areas with high populations (including the East Coast), while large sections of central U.S. lack air monitoring data.
Analyzing Relationships between PM2.5 and Mortality
Crude Model
Now the primary question is whether counties with higher PM2.5 also have higher mortality rates. To answer this question, first we need to perform some data merging in preparation for this analysis.
# Merging mortality and air pollution data
model_data <- merge(epa_ap_county, cdc_mortality, by.x = "State_County_Code", by.y = "County.Code")As we saw in the above plot, only a portion of the USA is covered by PM2.5 monitors. Let’s see what our population coverage is
sum(model_data$Population, na.rm = TRUE)
#> [1] 232169063
sum(cdc_mortality$Population, na.rm = TRUE)
#> [1] 323127513
sum(model_data$Population, na.rm = TRUE)/sum(cdc_mortality$Population, na.rm = TRUE)*100
#> [1] 71.8506We can do a quick visual inspection of this using a scatter plot which will also let us check for unexpected distributions of the data (always a good idea)
ggplot(model_data) +
geom_point(aes(x = Age.Adjusted.Rate, y = PM25)) +
ggtitle(expression(PM[2.5]~by~Mortality~Rate)) +
xlab('Age-adjusted Mortality Rate') + ylab(expression(PM[2.5])) # changing axis labels 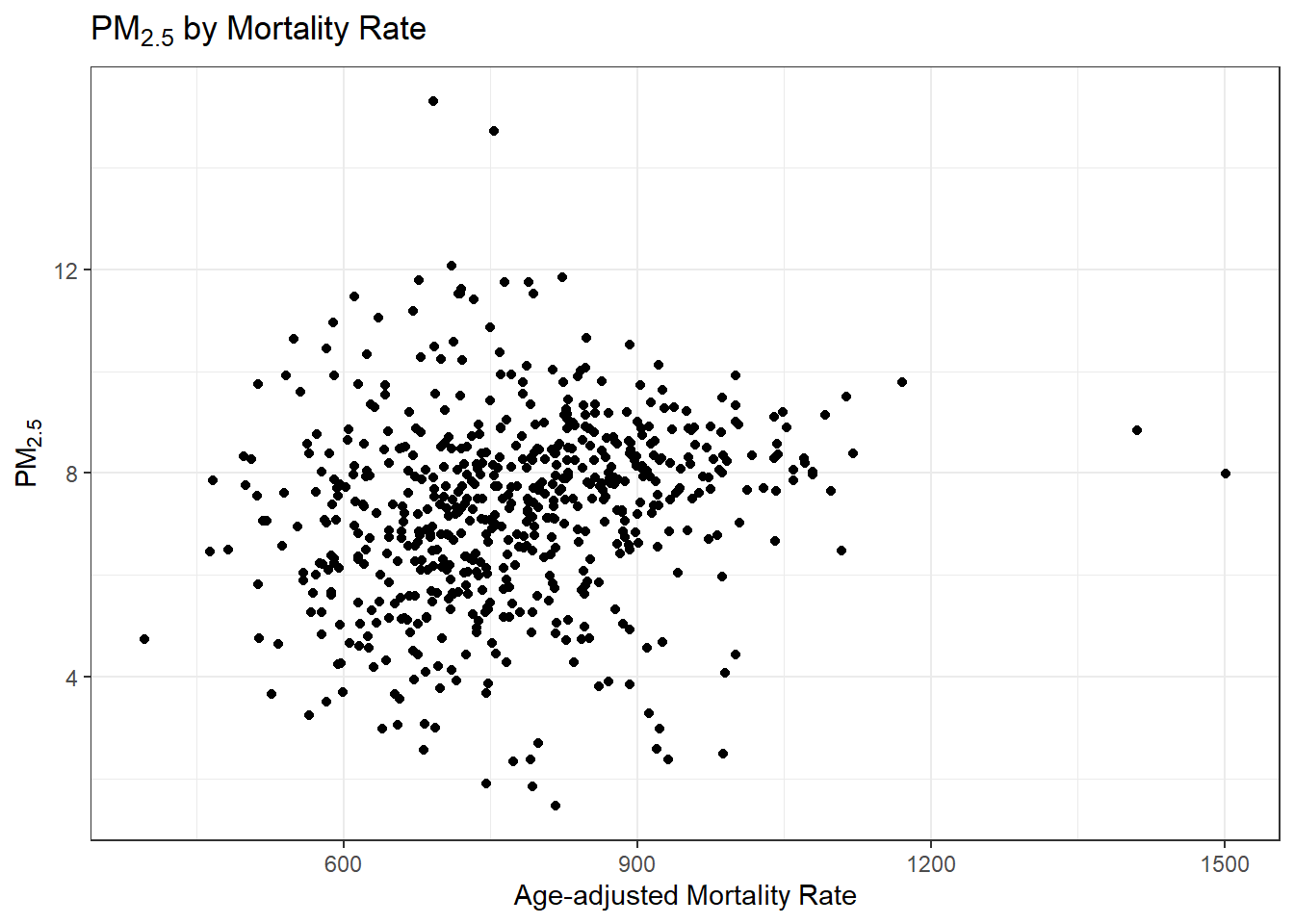
The univariate correlation is a simple way of quantifying this potential relationship, though it does not nearly tell the complete story. Just as a starting point, let’s run this simple univariate correlation calculation using the cor() function.
Now, let’s obtain a more complete estimate of the data through regression modeling. As an initial starting point, let’s run this model without any confounders (also known as a ‘crude’ model).
A simple linear regression model in R can be carried out using the lm() function. Here, we are evaluating age-adjusted mortality rate (age.adjusted.rate) as the dependent variable, and PM2.5 as the independent variable. Values used in evaluating this model were weighted to adjust for the fact that some counties have higher precision in their age-adjusted mortality rate (represented by a smaller age-adjusted rate standard error).
# running the linear regression model
m <- lm(Age.Adjusted.Rate ~ PM25,
data = model_data, weights = 1/model_data$Age.Adjusted.Rate.Standard.Error)
# viewing the model results through the summary function
summary(m)
#>
#> Call:
#> lm(formula = Age.Adjusted.Rate ~ PM25, data = model_data, weights = 1/model_data$Age.Adjusted.Rate.Standard.Error)
#>
#> Weighted Residuals:
#> Min 1Q Median 3Q Max
#> -110.096 -9.792 8.363 25.090 84.316
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 661.038 22.528 29.343 < 2e-16 ***
#> PM25 8.956 2.878 3.112 0.00195 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 29.93 on 608 degrees of freedom
#> (88 observations deleted due to missingness)
#> Multiple R-squared: 0.01568, Adjusted R-squared: 0.01406
#> F-statistic: 9.685 on 1 and 608 DF, p-value: 0.001945Shown here are summary level statistics summarizing the results of the linear regression model.
In the model summary we see several features:
Estimate: the regression coefficient which tells us the relationship between a 1 ug/m3 change (elevation) in PM2.5 and the age-adjusted all-cause mortality rateStd. Error: the standard error of the estimatet value: represents the T-statistic which is the test statistic for linear regression models and is simply theEstimatedivided byStd. Error. This t value is compared with the Student’s T distribution in order to determine the p-value (Pr(>|t|)).
The residuals are the difference between the predicted outcome (age-adjusted mortality rate) and known outcome from the data. For linear regression to be valid this should be normally distributed. The residuals from a linear model can be extracted using the residuals() function and plotted to see their distribution.
Answer to Envrionmental Health Question 2
With this, we can answer Environmental Health Question #2: Is there an association between long-term, ambient PM2.5 concentrations and mortality at the county level?
Answer: Based on these model results, there may indeed be an association between PM2.5 concentrations and mortality (p=0.0019)
Adjusting for Covariates
To more thoroughly examine the potential relationship between PM2.5 concentrations and mortality it is absolutely essential to adjust for confounders. Let’s start by viewing the human covariate data that contains some confounders of interest.
These covariate data are represented by the following columns:
State.FIPS.Code: Single digit code assigned to each stateCounty.FIPS.Code: FIPS code unique to counties within a particular state (1 digit)County.5.digit.FIPS.Code: Unique county FIPS code (5 digits)
State.Abbreviation: State abbreviationName: County Name (or state name that contains the state average)Release.Year: Year the data was publicly released- Various confounders indicative of race, ethnicity, socioeconomic status, population density, education, health-related variables in columns 7-36
# Merging the covariate data in with the AQS data
model_data <- merge(model_data, county_health, by.x = "State_County_Code",
by.y = "County.5.digit.FIPS.Code", all.x = TRUE)
# Now we add some relevant confounders to the linear regression model
m <- lm(Age.Adjusted.Rate ~ PM25 + Adult.smoking + Adult.obesity + Food.environment.index +
Physical.inactivity + High.school.graduation + Some.college + Unemployment +
Violent.crime + Percent.Rural + Percent.Females + Percent.Asian +
Percent.Non.Hispanic.African.American + Percent.American.Indian.and.Alaskan.Native +
Percent.NonHispanic.white, data = model_data, weights = 1/model_data$Age.Adjusted.Rate.Standard.Error)
# And finally we check to see if the statistical association persists
summary(m)
#>
#> Call:
#> lm(formula = Age.Adjusted.Rate ~ PM25 + Adult.smoking + Adult.obesity +
#> Food.environment.index + Physical.inactivity + High.school.graduation +
#> Some.college + Unemployment + Violent.crime + Percent.Rural +
#> Percent.Females + Percent.Asian + Percent.Non.Hispanic.African.American +
#> Percent.American.Indian.and.Alaskan.Native + Percent.NonHispanic.white,
#> data = model_data, weights = 1/model_data$Age.Adjusted.Rate.Standard.Error)
#>
#> Weighted Residuals:
#> Min 1Q Median 3Q Max
#> -70.059 -7.055 0.960 8.119 64.052
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 616.47976 157.20842 3.921 1.00e-04 ***
#> PM25 3.84851 1.69321 2.273 0.023444 *
#> Adult.smoking 859.27337 137.81312 6.235 9.43e-10 ***
#> Adult.obesity 605.84315 97.71952 6.200 1.16e-09 ***
#> Food.environment.index -28.65536 3.93359 -7.285 1.22e-12 ***
#> Physical.inactivity 117.60916 91.41519 1.287 0.198834
#> High.school.graduation 55.14446 40.78490 1.352 0.176944
#> Some.college -244.41255 49.78764 -4.909 1.23e-06 ***
#> Unemployment 97.98159 161.89025 0.605 0.545290
#> Violent.crime 0.07551 0.01516 4.982 8.62e-07 ***
#> Percent.Rural -12.55308 20.84834 -0.602 0.547364
#> Percent.Females -271.45504 299.16925 -0.907 0.364640
#> Percent.Asian 74.54327 55.89262 1.334 0.182897
#> Percent.Non.Hispanic.African.American 153.39910 39.49623 3.884 0.000116 ***
#> Percent.American.Indian.and.Alaskan.Native 200.70010 102.75028 1.953 0.051329 .
#> Percent.NonHispanic.white 240.17976 26.30538 9.130 < 2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 14.22 on 514 degrees of freedom
#> (168 observations deleted due to missingness)
#> Multiple R-squared: 0.7841, Adjusted R-squared: 0.7778
#> F-statistic: 124.4 on 15 and 514 DF, p-value: < 2.2e-16Answer to Envrionmental Health Question 3
With this, we can answer Environmental Health Question #3: What is the difference when running crude statistical models vs. statistical models that adjust for potential confounding, when evaluating the relationship between PM2.5 and mortality?
Answer: The relationship between PM2.5 and mortality remains statistically significant when confounders are considered (p=0.023), though is not as significant as when running the crude model (p=0.0019).
Environmental Justice Considerations
Environmental justice is the study of how societal inequities manifest in differences in environmental health risks either due to greater exposures or a worse health response to exposures. Racism and racial discrimination are major factors in both how much pollution people are exposed to as well what their responses might be due to other co-existing inequities (e.g. poverty, access to healthcare, food deserts). Race is a commonly used proxy for experiences of racism and racial discrimination.
Here we will consider the race category of Non-Hispanic African-American to investigate if pollution levels differ by percent African-Americans in a county and if associations between PM2.5 and mortality also differ by this variable, which could indicate environmental justice-relevant issues revealed by this data. We will specifically evaluate data distributions across counties with the highest percentage of African-Americans (top 25%) vs. lowest percentage of African-Americans (bottom 25%).
First let’s visualize the distribution of African-American percentage in these data
ggplot(model_data) +
geom_histogram(aes(x = Percent.Non.Hispanic.African.American*100)) +
ggtitle("Percent African-American by County") +
xlab('Percent')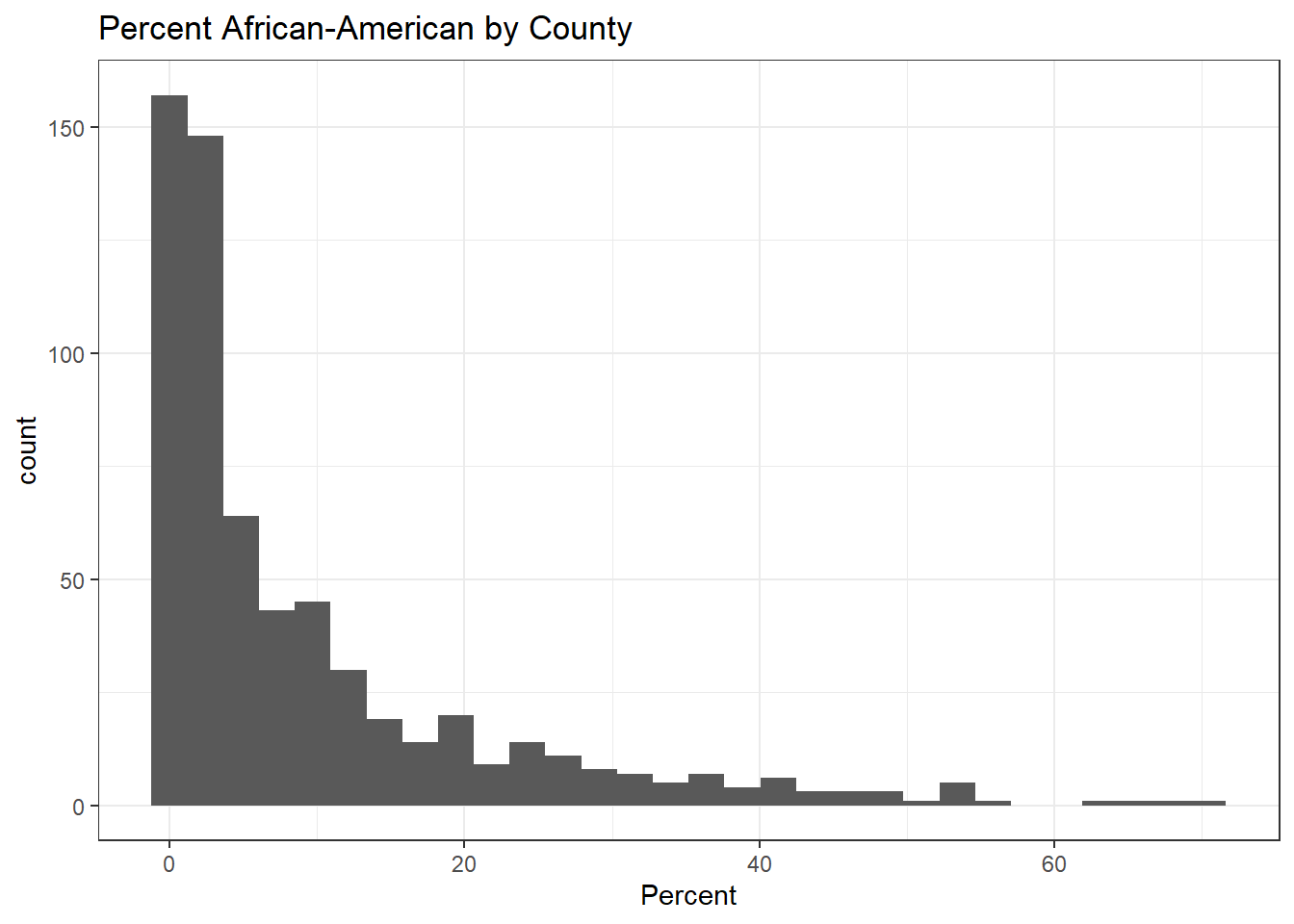
Let’s look at a summary of the data
summary(model_data$Percent.Non.Hispanic.African.American)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 0.00056 0.01220 0.03995 0.09415 0.12282 0.70458 67We can compute quartiles of the data
model_data$AA_quartile <- with(model_data, cut(Percent.Non.Hispanic.African.American,
breaks = quantile(Percent.Non.Hispanic.African.American, probs = seq(0,1, by = 0.25), na.rm = TRUE),
include.lowest = TRUE, ordered_result = TRUE, labels = FALSE))Then we can use these quartiles to see that as the Percent African-American increases so does the PM2.5 exposure by county
AA_summary <- model_data %>%
filter(!is.na(Percent.Non.Hispanic.African.American)) %>%
group_by(AA_quartile) %>%
summarise(Percent_AA = mean(Percent.Non.Hispanic.African.American, na.rm = TRUE), Mean_PM25 = mean(PM25, na.rm = TRUE))
AA_summary
#> # A tibble: 4 × 3
#> AA_quartile Percent_AA Mean_PM25
#> <int> <dbl> <dbl>
#> 1 1 0.00671 5.97
#> 2 2 0.0238 7.03
#> 3 3 0.0766 7.93
#> 4 4 0.269 8.05Now that we can see this trend, let’s add some statistics. Let’s specifically compare the relationships between PM2.5 and mortality within the bottom 25% AA counties (quartile 1); and also the highest 25% AA counties (quartile 4)
# first need to subset the data by these quartiles of interest
low_AA <- subset(model_data, AA_quartile == 1)
high_AA <- subset(model_data, AA_quartile == 4)
# then we can run the relevant statistical models
m.low <- lm(Age.Adjusted.Rate ~ PM25 + Adult.smoking + Adult.obesity + Food.environment.index + Physical.inactivity +
High.school.graduation + Some.college + Unemployment + Violent.crime + Percent.Rural + Percent.Females +
Percent.Asian + Percent.American.Indian.and.Alaskan.Native + Percent.NonHispanic.white,
data = low_AA, weights = 1/low_AA$Age.Adjusted.Rate.Standard.Error)
m.high <- lm(Age.Adjusted.Rate ~ PM25 + Adult.smoking + Adult.obesity + Food.environment.index + Physical.inactivity +
High.school.graduation + Some.college + Unemployment + Violent.crime + Percent.Rural + Percent.Females +
Percent.Asian + Percent.American.Indian.and.Alaskan.Native + Percent.NonHispanic.white,
data = high_AA, weights = 1/high_AA$Age.Adjusted.Rate.Standard.Error)
# We see a striking difference in the associations
rbind(c("Bottom 25% AA Counties",round(summary(m.low)$coefficients["PM25",c(1,2,4)],3)),
c("Top 25% AA Counties",round(summary(m.high)$coefficients["PM25",c(1,2,4)],3)))
#> Estimate Std. Error Pr(>|t|)
#> [1,] "Bottom 25% AA Counties" "4.782" "3.895" "0.222"
#> [2,] "Top 25% AA Counties" "14.552" "4.13" "0.001"Answer to Envrionmental Health Question 4
With this, we can answer Environmental Health Question #4: Do observed associations differ when comparing between counties with a higher vs. lower percentage of African-Americans which can indicate environmental justice concerns?
Answer: Yes. Counties with the highest percentage of African-Americans (top 25%) demonstrated a highly significant association between PM2.5 and age-adjusted mortality, even when adjusting for confounders (p=0.001), meaning that the association between PM2.5 and mortality within these counties may be exacerbated by factors relevant to race. Conversely, counties with the lowest percentages of African-Americans (bottom 25%) did not demonstrate a significant association between PM2.5 and age-adjusted mortality, indicating that these counties may have lower environmental health risks due to factors correlated with race.
Concluding Remarks
In conclusion, this training module serves as a novel example data integration effort of high relevance to environmental health issues. Databases that were evaluated here span exposure data (i.e., Air Quality System data), health outcome data (i.e., mortality data), and county-level characteristics on healthcare, food environment, and other potentially relevant confounders (i.e., county-level variables that may impact observed relationships), and environmental justice data (e.g., race). Many different visualization and statistical approaches were used, largely based on linear regression modeling and county-level characteristic stratifications. These example statistics clearly support the now-established relationship between PM2.5 concentrations in the air and mortality. Importantly, these related methods can be tailored to address new questions to increase our understanding between exposures to chemicals in the environment and adverse health outcomes, as well as the impact of different individual or area characteristics on these relationships - particularly those that might relate to environmental justice concerns.
This training module provided some examples looking at PM2.5 concentration data. Using the same “Module7_4_InputData.data” file (also saved as “Module7_4_TYKInput.data”), let’s ask similar questions but now looking at NO2 concentration data.
- Is there an association between long-term, ambient NO2 concentrations and age-adjusted mortality at the county level (i.e., the crude model results)?
- After adjusting for covariates, is there an association between long-term, ambient NO2 concentrations and age-adjusted mortality at the county level (i.e., the adjusted model results)?